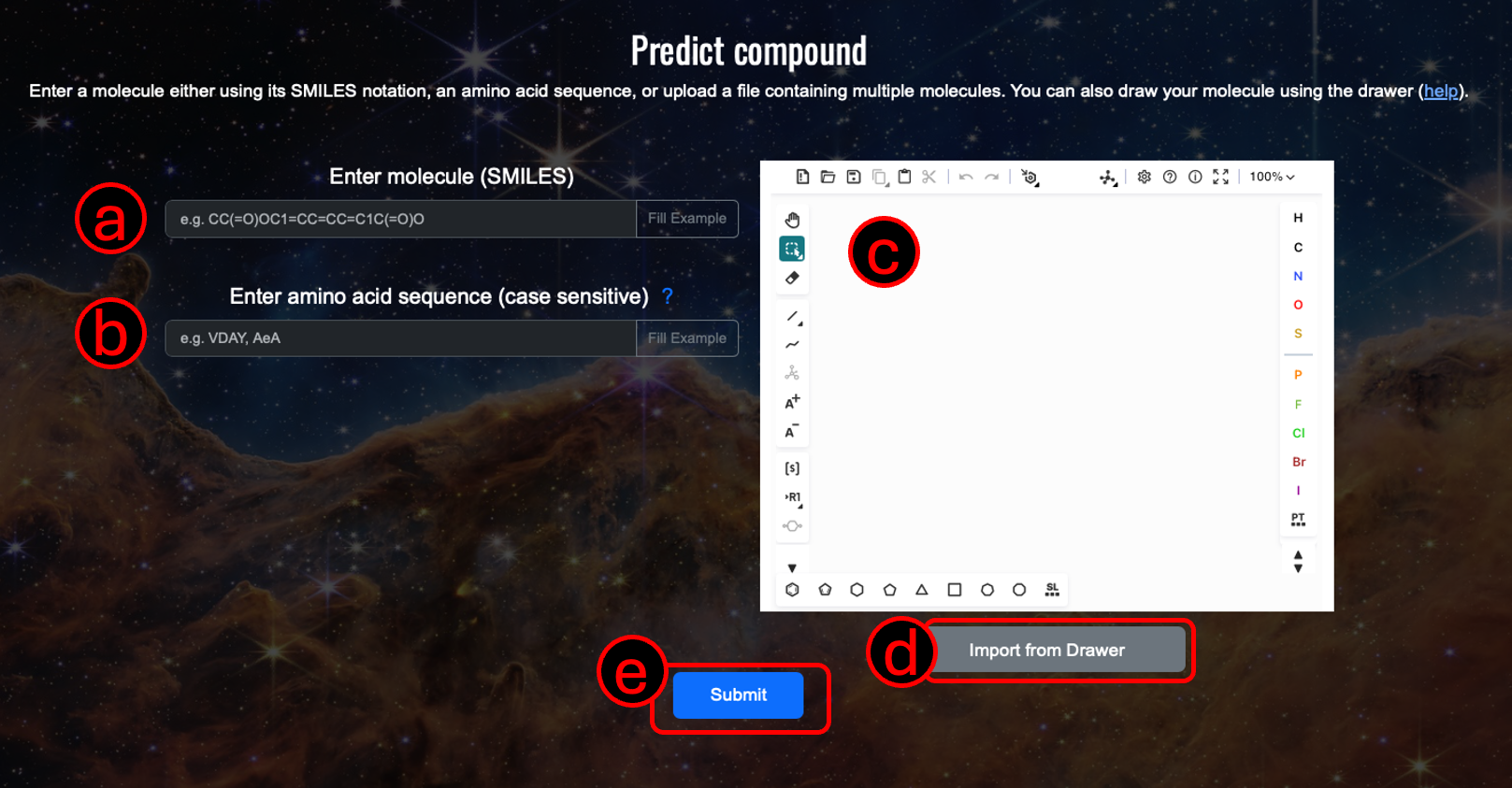
How to use the Predict page:
1. Input a molecule using (a) its SMILES notation or (b) amino acid sequence
IMPORTANT: this field is case sensitive, use single-letter amino acid codes, UPPERCASE for L-amino acid and lowercase for D-amino acid, you can also mix them (e.g., AAeEEG)
Optional: Use (c) the molecule drawer to draw your molecule and click (d) to export its SMILES notation to input above.
2. Click the "Predict" button (e) to start the prediction process.
3. You will be redirected to the wait page, a uniquie link will be generated for your job submission. Wait for the results to be generated and get redirected on the results page.
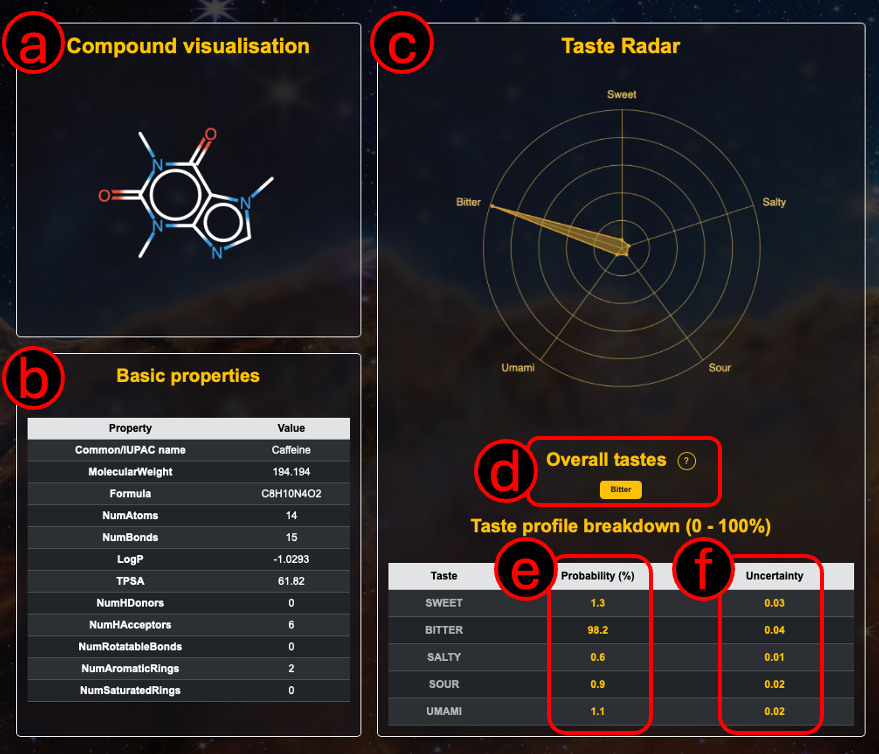
How to interpret the Results page:
The results page displays the predicted taste profiles for the submitted molecule. Each taste category (e.g., sweet, bitter, umami) is represented with a probability score ranging from 0 to 1.
In the page you can find:
a. The visualised molecule (note that we use rdkit to canonicalise the input molecule, so the 2D structure may differ from your input), but nevertheless it is the same compound
b. Basic properties of the molecule such as molecular weight, number of atoms, number of bonds, etc.
c. A radar chart showing the predicted probabilities for each basic taste
d. Tastes that are above the 50% threshold
e. Probabilities for each basic taste, same as in (c) the radar chart
f. Uncertainty quantification for each taste prediction (lower is better)
For all web server related issues please contact Kenny Lam directly at: plete2026 [at] gmail.com